What is extrachromosomal DNA? How have we come to think of its role in cancer? And what has all this got to do with a flagship lung cancer study? Here, Dr Chris Bailey tell us how ecDNA challenges traditional models of tumour evolution and drug resistance and why TRACERx has been pivotal in pushing forward our understanding of this mysterious molecule…
The idea that Darwinian principles might play a role in driving different cancer cell populations in the same tumour was put forward in the late 1970’s by Peter Nowell. The concept is one where cancer cells – under the principles of variation, heritability of fitness and selection – compete to survive microenvironmental pressures.
These pressures include hypoxia, stroma, the immune system, and therapies. It is through this mechanism of evolution that some tumours can ultimately become resistant to therapy. Variation comes in the form of intratumour heterogeneity, and this is often driven by underlying genomic instability.
“It’s clear this heterogeneity is very important clinically, which is why understanding extrachromosomal DNA (ecDNA) could be so useful.”
So, it’s clear this heterogeneity is very important clinically, which is why understanding extrachromosomal DNA (ecDNA) could be so useful – it offers an extreme evolutionary path to heterogeneity and resistance because it allows tumours to rapidly amplify multiple copies of oncogenes. And, over the last 10 years, key discoveries in the field have indeed highlighted the role of ecDNA in tumour evolution and therapy resistance.
What is extrachromosomal DNA?
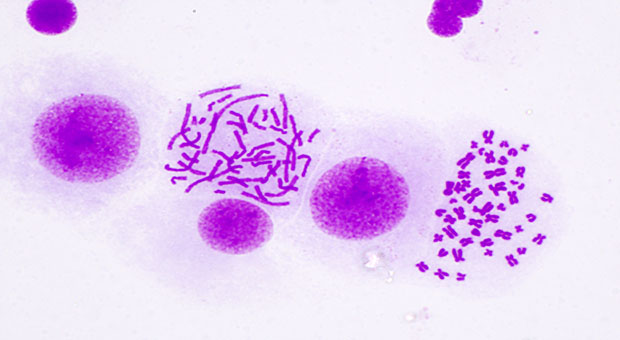
ecDNAs are circular DNA structures ranging from hundreds of kilobases to several megabases and often contain genes and regulatory elements that drive tumour growth and proliferation. They lack centromeres and are subject to random segregation during mitosis and can be present at hundreds of copies within a single cell. ecDNA has been shown to drive high levels of expression through co-operative hubs. These are curious structures that form when tens – or even hundreds – of individual free ecDNA molecules cluster together. It is also known that reintegration of ecDNA into a chromosome to form homogenous staining regions (HSR) have been shown to contribute to extreme intrachromosomal gene amplification.
There are several routes to ecDNA formation. Perhaps the best described is chromothripsis – a catastrophic mutational event that results in multiple chromosomal rearrangements – something that can affect multiple chromosomes. ecDNA has also been described following an episome formation – structures that arise when genomic fragments are excised from a chromosome and subsequently take a circular form.
Historically, ecDNA has been described on cytogenetic preparations as ‘double minutes’ (DMs) due to their paired structure. The term HSR was coined over a decade later due the homogenous appearance of the structure seen when karyotyping chromosomes.
Circular DNA structures are not unique to cancer cells. Together, they are commonly classified as extrachromosomal circular DNA (eccDNA) and include small polydispersed DNA, telomeric circles and microDNA. eccDNA are commonly found in yeast such as S. cerevisiae and implicated in glycophosphate resistance in A. palmeri. There are also other extrachromosomal elements in nature, such as plasmids, viruses, and, most recently described, Borgs. These, rather incredibly, are genetic elements of up to 1 megabase in length that seem to have originated from methane-oxidizing Methanoperedens archaea.
ecDNA and cancer
The development of high throughput sequencing and bioinformatic methods to detect and quantify ecDNA has allowed the creation of large datasets derived from patient samples, and this has begun to help elucidate exactly what impact they have on tumour biology.
In 2020, Roel Verhaak and colleagues published an analysis involving over 3,200 patient samples. They showed that ecDNA was associated with aggressive tumour behaviour and poorer survival outcomes in over 14% of samples analysed and in 25 of the 29 tumour types – most frequently occurring in glioblastoma, oesophageal carcinoma and sarcomas.
“We really need to better understand the mechanisms by which ecDNA is generated, whether non-tumour cells can produce ecDNA under stress and what the relationship is between ecDNA and the immune system.”
ecDNA has also been implicated in targeted therapy and chemoresistance. In 1981, Daniel Haber and Robert Schimke reported that methotrexate resistance can occur due to di-hydrofolate reductase amplification on ecDNA. This phenomenon was recently shown to occur due to chromothripsis in response to increasing therapeutic pressure in HeLa cells. Then, in 2014, Paul Mischel demonstrated just how import that ability of ecDNA to reincorporate into a chromosome as homogenous staining regions could be. His team showed that, in glioblastoma, tumour cells with EGFRvIII-amplified ecDNA treated with an EGFR tyrosine kinase inhibitor became resistant through the dynamic reintegration of ecDNA as HSRs. Intriguingly, on removal of treatment, the ecDNA re-emerged. This reintegration of ecDNA back into chromosomes has also been shown to inactivate tumour suppressor genes.
There have been several emerging methods to detect, isolate and model ecDNA that infer ecDNA through genome sequencing data, as well as traditional cytogenetic methods such as giemsa-banding and fluorescence in situ hybridisation. One method, CRIPSR-CATCH, utilises the fact that ecDNA, when linearised through CRISPR mediated ‘cuts’, can be separated out using electrophoresis.
With the development of techniques like this, the field is better placed to establish models to answer some basic unknowns. We really need to better understand the mechanisms by which ecDNA is generated, whether non-tumour cells can produce ecDNA under stress and what the relationship is between ecDNA and the response of the immune system. Making progress on these unknowns may have real implications for clinical application.
TRACERx and ecDNA
The TRACERx and PEACE studies have provided an excellent opportunity to study ecDNA biology in patients.
TRACERx is a multi-site, longitudinal study that set out to understand the association between intra-tumour heterogeneity and clinical outcome and has made several key discoveries in lung cancer evolution. PEACE, closely linked with TRACERx, is a national autopsy study where patients consent to sampling of metastatic disease post-mortem. ecDNA is currently estimated to be present in just under 25% of patients with non-small cell lung cancer. By sampling cancers at surgery, after relapse and at autopsy, we are in an excellent position to begin to understand how ecDNA evolves through treatment. In this study, whole genome sequencing data is used to infer ecDNA through computational software. This is combined with RNA or DNA in situ hybridisation for direct visualisation.
ecDNA is also the topic of a Cancer Grand Challenge, which has provided a unique opportunity to generate a collaborative, multidisciplinary programme to accelerate discoveries in this understudied phenomenon.
Through diverse approaches from fields as diverse as cancer genomics and chemical biology to mathematical modelling and immunology, further understanding of the role ecDNA plays in tumour biology and clinical pathogenesis may inform novel treatment strategies for cancer patients.
 Dr Chris Bailey is a Specialist Registrar in Haematology and Clinical Research Fellow at The Francis Crick Institute
Dr Chris Bailey is a Specialist Registrar in Haematology and Clinical Research Fellow at The Francis Crick Institute
Tags
There are no tags