February 12, 2019, by NCI Staff
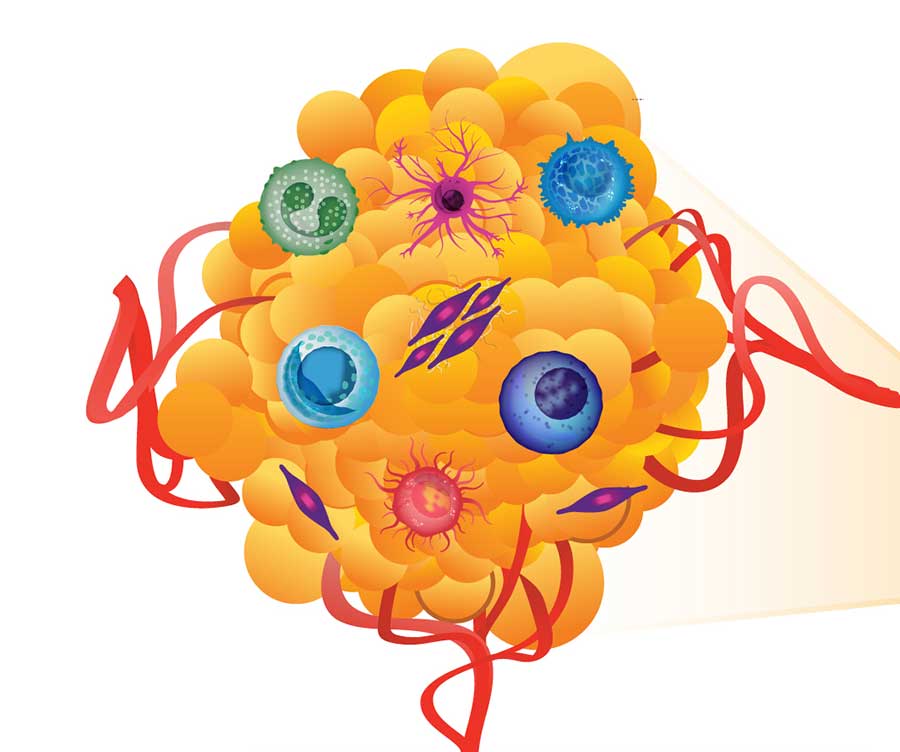
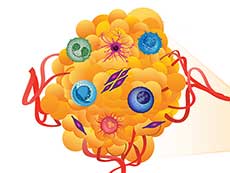
The Human Tumor Atlas Network initiative will create maps of human tumors, including their surrounding environment, which can include immune cells, blood vessels, and other components of the tumor’s supporting structure.
Credit: National Cancer Institute
If you’re heading to a completely new destination, chances are you might consult a website or app that predicts the best route using up-to-date maps and traffic information.
What if oncologists had a similar resource that used highly detailed maps of cancer to predict the best route to treat someone with cancer or prevent cancer in someone who is at increased risk?
The Human Tumor Atlas Network (HTAN), an NCI-funded collaborative research project, was established to do just that—create detailed “maps” of a variety of cancers that may be used to investigate how cancer might develop, spread, or respond to treatment.
The maps will capture the molecular features of cancer, similar to what has been done with other cancer “atlases.” However, the most unique aspect of HTAN is that the features of cancer will be mapped across space and time.
In addition, HTAN researchers will leverage the latest biotechnologies and computational approaches to study the biology of cancer in unprecedented ways.
For example, the HTAN maps will incorporate the 3-dimensional location of individual cells in a tumor, like building addresses. And just like an interactive map that indicates whether a building is a home or a business, the HTAN maps will also be annotated with information about the type of cell at each “address.”
But tumors, like cities, change over time, and an out-of-date map is not a very reliable resource. As such, the HTAN maps will depict how the molecular, cellular, and spatial features of a tumor change as it progresses over time.
With these highly detailed maps, HTAN aims to generate a more comprehensive understanding of cancer than was previously possible, the initiative’s leaders said.
“A Very Timely Effort”
The idea for HTAN came from the Cancer Moonshot℠, in the form of a recommendation from its Blue Ribbon Panel.
“It is a very timely effort because there has been a real blossoming of the technologies that are required to look at cells in depth, to an extent that was not possible 10 years ago,” said HTAN co-leader Tracy Lively, Ph.D., of NCI’s Division of Cancer Treatment and Diagnosis.
And these technologies have recently become available “not just to a few specialized labs, but broadly enough that labs with deep interest and expertise in cancer can now harness them,” she added.
The network consists of 10 teams from research institutions across the country. At a kickoff meeting held last November, each team outlined how it is generating tumor atlases using a different combination of technologies and approaches.
“When we are trying to understand the underlying biology [of cancer], we really need a highly multifaceted approach and to approach it from many different angles,” said Andrew Adey, Ph.D., an HTAN-funded scientist from Oregon Health and Science University.
Each map will represent a particular type of cancer as it goes through a specific transition. For example, the HTAN team from Children’s Hospital of Philadelphia is generating a map of pediatric high-grade glioma as it transitions from responding to standard treatments to developing resistance to them.
Getting Down to Street Level: Analyzing Single Cells
Looking at a city with a birds-eye view can reveal general structures like buildings and parks. But with a street-level view, you can see individual details of those places, such as whether a building is residential or commercial.
In the past, it was difficult for researchers to get such a “street-level view” of cells because, for most laboratory tests, a sample of cells is mixed together and analyzed as a group.
It’s like putting a bunch of cells in a blender and looking at the result, said Dana Pe’er, Ph.D., a leader of the HTAN team at Memorial Sloan Kettering Cancer Center (MSKCC). With this “bulk” approach, Dr. Pe’er explained, the individual differences (called heterogeneity) are lost.
However, technological advances now allow researchers to quickly and affordably analyze molecular characteristics of single cells. By combining individual data from hundreds or thousands of single cells from a tumor, researchers can get a better handle on its heterogeneity.
These single-cell technologies are “opening an entire new world for cancer and biomedicine,” Dr. Pe’er said.
Each HTAN team is using different single-cell technologies, but their end-goal is the same: to map the location of and understand the role of each type of cell in a tumor. For example, the researchers hope to differentiate cancer cells and immune cells, and to determine if those immune cells are helping the cancer grow or attacking it.
“To understand not just the tumor cells, but also the surrounding microenvironment, we need to utilize newer technologies … to really dissect the different populations in a tumor,” said Li Ding, Ph.D., a leader of the team at Washington University in St. Louis.
Looking Beyond the DNA Blueprint
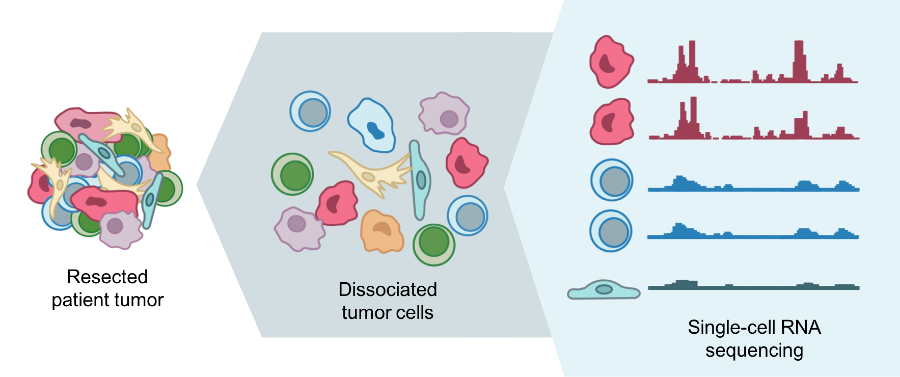
Single-cell RNA sequencing can help identify different types of cells that are present in a tumor.
Credit: Mario Suva, M.D., Ph.D., Harvard Medical School, and Itay Tirosh, Ph.D., Broad Institute
A city’s blueprint might show you the original outline of streets and highways, but it couldn’t tell you what buildings and landscapes have popped up along those streets, or how the outline was altered as the city was developed.
Similarly, while there’s no doubt that the sequence of DNA (often referred to as the blueprint of a cell) reveals clues about cancer biology, it’s not the whole story.
In addition to DNA sequencing, HTAN researchers are using single-cell technologies to study other biological molecules such as RNA and proteins, which can reveal important information about cancer cells.
For example, the MSKCC team is studying the dynamics of metastasis by sequencing RNA of single cells from primary tumors in the lung and pancreas, and metastatic tumors in the brain.
With single-cell RNA sequencing, the researchers can identify what type of cell it is and determine its traits, such as the potential to metastasize, Dr. Pe’er said. With their maps, they hope to gain a better understanding of the mechanisms of metastasis and how they are regulated.
Other teams are studying how epigenetic modifications—alterations that control which genes are switched on and off without changing the actual DNA sequence—shift as cancer progresses.
The team from Oregon Health and Science University, for example, is using a technology known as ATAC-seq to analyze one kind of epigenetic modification. This technology is more efficient and easier to use than previous epigenetic tests, explained Oregon team member Dr. Adey.
He and his colleagues adapted ATAC-seq to analyze single cells and are using this approach to study epigenetic changes that occur as metastatic breast tumors become resistant to standard therapies.
Multiple studies have shown that certain epigenetic shifts make cancer cells resistant to a given treatment. The team hopes that delving deeper into these shifts will help them better understand these resistance mechanisms and, possibly, how to subvert them.
To create their maps, the HTAN team from Washington University is using a new technology called nanoPOTS to identify hundreds of proteins in single cells. By analyzing the proteins in cells collected from breast, brain, and pancreatic tumors during and after treatment, the team hopes to learn how changes in the types of cells in a tumor correlate with the patient’s response to treatment.
For example, this approach could shed light on how the types of immune cells in a tumor change during and after treatment, explained Dr. Ding.
The hope is that, down the road, researchers may be able to use the HTAN atlases as a resource to probe for answers about cancer biology.
One way that researchers may utilize the maps, for example, is to “identify biomarkers for risk stratification and early cancer detection, and identify precise targets for prevention and therapy,” said HTAN co-leader Sudhir Srivastava, Ph.D., of NCI’s Division of Cancer Prevention.
Such markers and targets may enable oncologists to create models that predict how an individual’s cancer may progress and respond to treatment, he explained.
Studying Cancer in Context
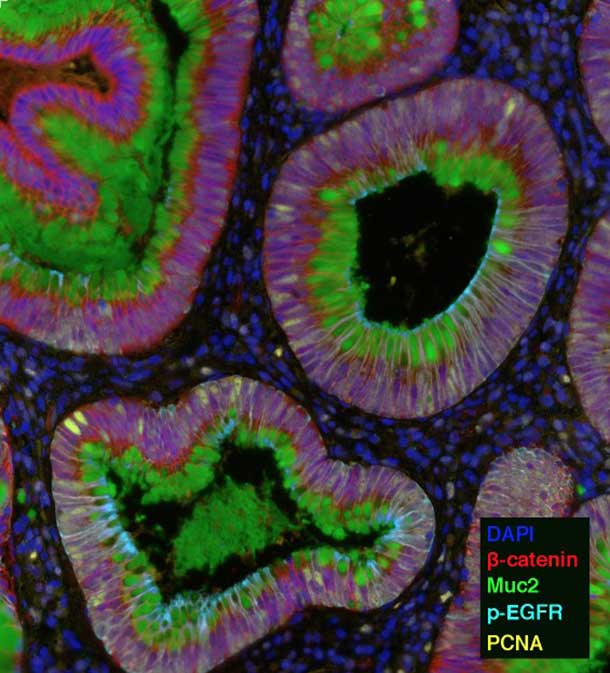
A sample of a human colon tumor analyzed by MxIF. Each of the five colors represents a different protein.
Credit: Eliot McKinley, Ph.D., Vanderbilt University School of Medicine
A map wouldn’t be able to direct you to a destination if the components of the city were drawn arbitrarily on the page. You’d need to know how the streets and buildings are organized in relation to one another to reach your destination.
Likewise, a tumor atlas needs to convey how the components of the tumor microenvironment—blood vessels, connective tissue, immune cells, and microorganisms—are organized in relation to one another and to cancer cells.
But in the past, said Dr. Lively, “many of our technologies for looking at cells required us to take the tumor apart,” thereby losing the spatial relationships between these components.
Advanced imaging technologies now enable researchers to study the intact tumor microenvironment in great detail. Using such technologies to tie molecular and spatial information together is a major objective of HTAN, Dr. Lively noted.
These methods “are incredibly important because you can study the cells in context,” said Dr. Pe’er. For example, one could address a question like, “how does a cancer cell next to an immune cell behave differently than one that isn’t?” she said.
HTAN investigators are using several imaging techniques that can single out different molecules—sometimes upwards of 100—by highlighting each one with a different color.
For example, the HTAN team from Vanderbilt University is using one technique, called multiplex immunofluorescence (MxIF), that can highlight up to 60 different molecules, including proteins, RNA, and microbes. They are using this technology to study how the microenvironment changes during colon cancer development.
“Imaging-based approaches like multiplex immunofluorescence are going to be key for these efforts,” said Ken Lau, Ph.D., a leader of the Vanderbilt team.
The Vanderbilt researchers will interrogate combinations of proteins and RNA to delineate the types of cells in samples of precancerous growths in the colon and full-blown colon tumors. They will then analyze how the interaction and organization of these cells changes during cancer progression.
The team is also using MxIF to study microorganisms, because certain bacterial species in the gut are linked to the development and progression of colon cancer.
“The microbiome organizes into structures called biofilms,” Dr. Lau explained. “It’s very important to look at how these biofilms are spatially organized and where exactly the different microbes are,” he said. For instance, he said, microbes can sit on top of a colon tumor or they can invade into the tumor, and that can change how the tumor behaves.
Collaborating to Address New Challenges
The HTAN project is a large undertaking and the first 3-dimensional atlas project to focus solely on tumors, Dr. Lively noted. That means there will be a lot of kinks to work out, many of which the HTAN scientists discussed at the kickoff meeting.
One challenge will be isolating enough single cells for single-cell assays, Dr. Adey said. Sometimes large parts of a tumor sample have died and broken open, making it difficult to isolate single cells, he explained.
And even once that challenge is met, some of the newer technologies still need improvements, said Dr. Pe’er. At the meeting, the HTAN researchers agreed on the need to optimize each assay or test before it can be used on large sets of samples.
Computation and interpretation of large data sets is another big bottleneck, Dr. Pe’er said. Part of the issue is not having enough computational biologists, she noted.
Another challenge is generating datasets in a consistent way that allows them to be compared to one another, said Dr. Ding.
“We are dealing with a collection of different technologies and we need to be conscious about standardization. If we don’t, it will be difficult to make conclusions,” she said.
Although these are significant challenges, the combined expertise of the network is a good starting point to tackle these issues, Dr. Adey said. “Having this network of peers to exchange ideas and experiences with is going to be incredibly valuable,” he explained.
Dr. Lively agreed, saying, “I think the real value of putting the network together is that … there is going to be tremendous sharing and cross-fertilization.”