October 10, 2018, by NCI Staff
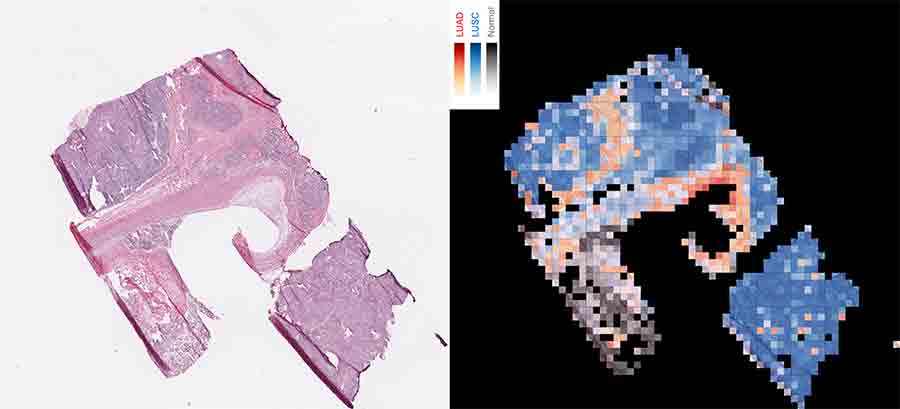
The AI-driven computer program analyzes tissue by creating a map of thousands of tiles. The map on the right shows squamous cell carcinoma (red), lung squamous cell carcinoma (blue), and normal lung tissue (gray).
Credit: NYU School of Medicine
Researchers have trained a computer program to read slides of tissue samples to diagnose two of the most common types of lung cancer with 97% accuracy. The program also learned to detect cancer-related genetic mutations in the samples just by analyzing the images of cancer tissue.
In a process known as machine learning, the computer program scanned images of tissue slices and developed the ability to differentiate normal lung tissue from the two most common forms of lung cancer, adenocarcinomas, which make up about 40% of lung cancers, and squamous cell carcinomas, which make up about 25% to 30% of lung cancers. Even experienced pathologists can struggle to distinguish these two types of lung cancer, which arise from different types of cells and require very different treatment regimens.
To train the computer program, researchers specializing in machine learning used a deep learning method originally developed and published by Google. The program uses artificial intelligence (AI) to teach itself to get better at a task—in this case, classifying lung cancer specimens—without being told exactly how.
The program was trained using more than 1,600 histopathology slides of lung specimens made publicly available by The Cancer Genome Atlas (TCGA). The study, led by researchers at New York University’s Langone Medical Center and published September 17 in Nature Medicine, represents a large improvement in the accuracy of computational methods to diagnose lung cancer; the second most accurate computational method has an accuracy rate of 83%.
Images as Data and a Public Resource
TCGA made histopathology images of tumor specimens available as a quality control measure for researchers studying the genetic sequence data collected in the project. The images “were required to make sure that the quality and identity of the tissue were correct,” said Jean C. Zenklusen, Ph.D., the director of TCGA at NCI. As an incidental benefit, Dr. Zenklusen said, the images themselves now serve as a resource for analysis.
The images made available by TCGA are large and high resolution, so the researchers at NYU divided each image into thousands of tiles, or “patches,” in a grid for the computer program to analyze individually for visual cues that could be linked to the sample’s classification. “We had about 500 patients per [lung cancer] subtype, but thousands of patches per image, so we had nearly one million patches in the end to train the model,” said Narges Razavian, Ph.D., a machine learning and AI researcher at NYU Langone, who helped lead the study.
The accuracy with which the program learned to distinguish adenocarcinoma from squamous cell carcinoma and normal lung cells was about equal to that of experienced pathologists, but the analysis can be done much faster; the program was able to reach a conclusion in a matter of seconds rather than the minutes a pathologist would need.
The program also correctly classified 45 of 54 images that at least one of three pathologists participating in the study misclassified, suggesting that AI could offer a useful second opinion, the researchers wrote.
The program was tested on an independent set of lung cancer specimens—both frozen and freshly collected—from NYU to verify that it worked on a completely separate collection of samples.
The samples from TCGA were almost entirely tumor tissue. In this validation set, however, the samples often included other components, such as blood clots and dead tissue, “making the classification task more challenging” for the program, the researchers reported.
In response, they reworked the program to have it focus on the parts of the tissue samples that were largely tumor (as identified by a pathologist). With that change, the method’s accuracy averaged more than 90%, which they described as “very encouraging.”
AI’s Role in Treatment and Research
Because of the computer program’s speed and accuracy, the research team suggested the tool could be used during surgery, for example, to help verify that a biopsy is of sufficient quality to make a diagnosis or to inform a surgeon that another sample is needed.
In addition to showing that AI can be used to make a quick and accurate diagnosis, the research project showed that AI could be trained to predict the presence of six of the most common genetic mutations in lung cancer adenocarcinoma, with an accuracy ranging from 64% to 86%, depending on the gene.
“That is very exciting scientifically,” Dr. Razavian said.
Currently, the only way humans can detect genetic mutations is by DNA sequencing, which can take up to 2 weeks.
“Lung cancer is usually detected late in the progression of the disease, so waiting 2 weeks to start any treatment is bad for the patient,” Dr. Razavian noted.
Many medical teams will start treatment and then adjust the medication used based on genetic test results. “What we show is that, with this program, you could start a treatment that is more likely to be the right one immediately,” Dr. Razavian said.
The program is a “black box,” in that its decisions are the result of thousands of interconnected small steps, not easy to summarize. The researchers can’t visualize exactly what the program detects in an image to predict the presence of genetic mutations, but the program can serve as “a lens that shows patterns that are hard to notice with the eye,” Dr. Razavian said.
She and her colleagues are using the program to research how genetic mutations affect the structures of cells and tissues. To approach this problem, they are using automated methods that modify the images to figure out what visual elements most influence the program’s ability to perceive mutations.
“The strengths of the study,” said Paula Jacobs, Ph.D., the associate director of NCI’s Cancer Imaging Program, “are the careful selection of the clinical problem to be addressed,” how the program was trained, and the fact that the results were verified using an independent set of lung cancer specimens.
Michael Snyder, Ph.D., chair of genetics at Stanford University, sees AI as the future of diagnosis. “I think we need to shift to using machine learning rather than rely on pathologists alone to do all the work,” he said. “Algorithms won’t replace pathologists, but they will assist them in making classifications. They will reduce the errors that pathologists would otherwise make.”
The code developed for the project is available for other researchers to use for other diagnostic applications. The NYU team has already started applying the code to learn to diagnose kidney, breast, and other cancers.