, by NCI Staff
Proteins are often called the workhorses of cells, the machinery that makes cells go. They act as key players in the many chemical reactions constantly occurring in cells, collect cellular garbage, provide structure, repair damaged DNA, and much more. But individual proteins rarely act on their own. Instead, like molecular LEGO bricks, they join together with other proteins in precise configurations to carry out their duties.
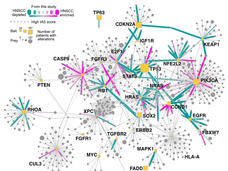
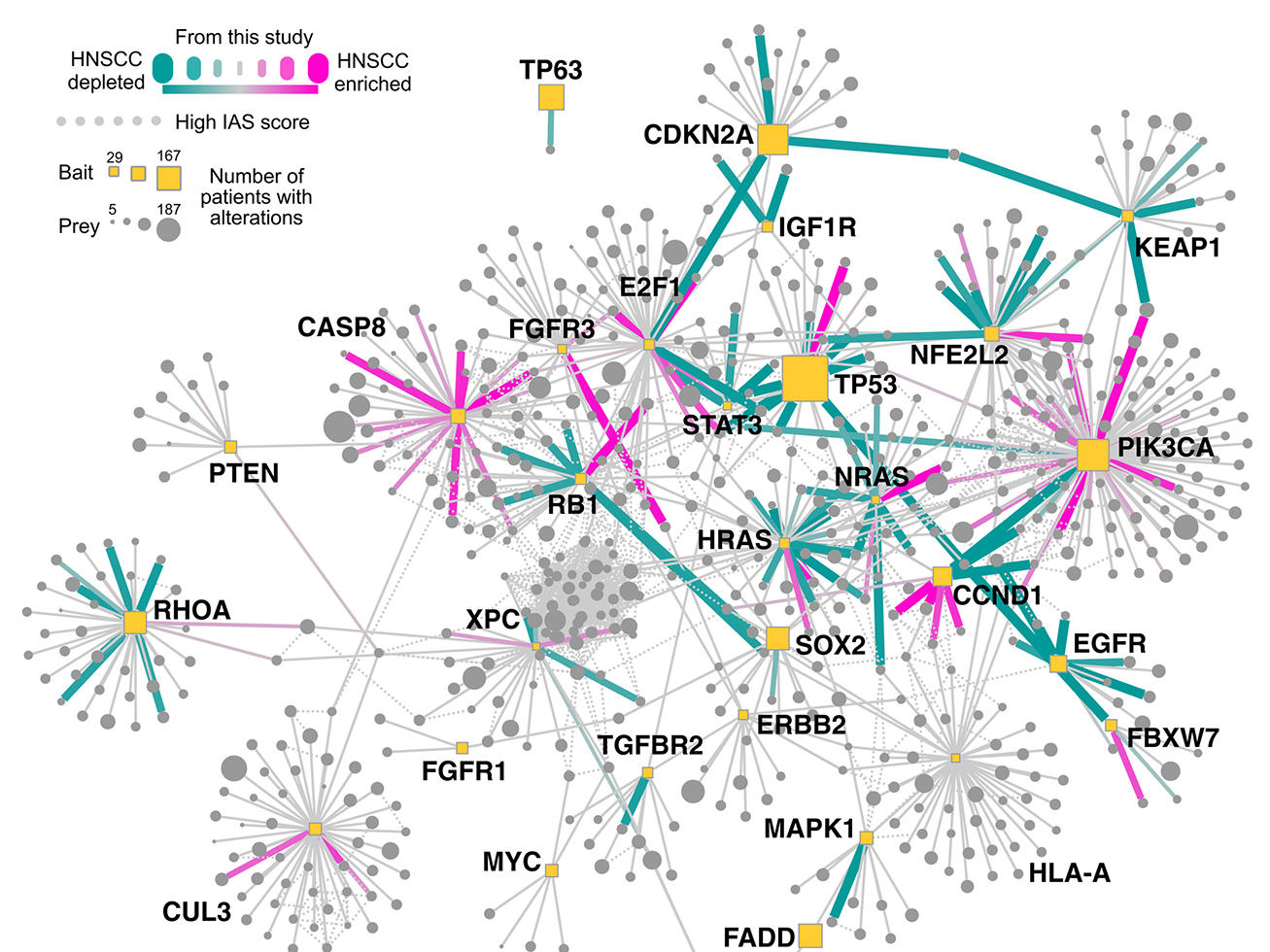
Now, a team of researchers has used advanced technologies and computational techniques to identify the interactions among proteins that commonly form in head and neck and breast cancer cells. They also created a map of how the interacting proteins—also called protein complexes—function in key communication networks, or pathways, in those cells.
Three studies describing the initial fruits of this work were published September 30 in Science.
In one of the studies, mapping protein–protein interactions in this fashion allowed them to identify a potentially effective treatment for people with head and neck cancers whose tumors have specific mutations in a gene called PIK3CA.
But unlike most targeted therapies, the drug doesn’t block the altered PIK3CA proteins in cancer cells that have mutations in the PIK3CA gene. Rather, the drug blocks the action of a different protein in the cell, called HER3.
The pathway map the team created revealed a strong interaction between the mutated PIK3CA proteins—which are part of a larger protein complex called PI3K—and HER3 proteins. Disrupting HER3, the researchers showed, provides a backdoor way of targeting the cancer-promoting actions of mutant PIK3CA.
This discovery, and important findings from the other two studies, provide some of the first true demonstrations of the potential of systematically mapping protein communication pathways in cancer cells, said one of the study leaders, Nevan Krogan, Ph.D., director of the Quantitative Biosciences Institute at the University of California, San Francisco (UCSF).
The effort builds on the many studies that have mapped cancer genomes while also addressing some of the shortcomings of gene-centric approaches to studying the biology of cancer, Dr. Krogan said.
“We need an approach that addresses the complexity [of cancer]. So we have to go to that next step,” he explained. That next step “is the functional unit of the cell: proteins.”
The studies are part of a larger NCI-supported research initiative by researchers at UCSF and the University of California San Diego (UCSD) to comprehensively map protein networks in cancer cells. The researchers believe these maps will open up new avenues for understanding and treating cancer.
At its core, the mapping effort is intended to provide “a higher-level view of cancer processes,” said Shannon Hughes, Ph.D., of NCI’s Division of Cancer Biology. “It’s not just focusing on one gene or one protein, but how [genes and proteins] act in a system.”
This line of research can provide important new insights into the biology of cancer, but also has the potential to impact cancer care, Dr. Hughes explained. “It broadens the potential targets for treatment,” she said. “And by taking that broader view, you can identify more patients who might be candidates for those treatments.”
From Genetic Mutations to Pathways
People with cancer are increasingly treated based on the genetic alterations present in their tumors. For example, several drugs are approved to treat patients with breast cancer and leukemia whose cancers have mutations in genes that provide the blueprints for the proteins that are part of the PI3K complex.
An ever-expanding collection of targeted cancer therapies block the actions of proteins produced as a result of mutations in genes like EGFR, HER2, BRAF, and many others. But with a few exceptions, most of these treatments only work in a modest portion of patients whose tumors express that target. Or, if they do initially shrink or stabilize tumors, in many cases the cancers become resistant to the treatment and begin growing again.
Researchers believe one reason these treatments don’t work or stop working is that specific alterations in cancer-related genes—even those that studies have shown are major “drivers” of a specific cancer—don’t represent the complete picture of how those mutations affect a cell’s protein machinery, explained the studies’ co-leader, Trey Ideker, Ph.D., a professor in the UCSD departments of medicine, bioengineering, and computer science.
Although it has had tremendous value, the more traditional gene-centric approach to studying cancer “is like looking at the nuts and bolts in a car engine,” Dr. Ideker said. “We’ve now got to look at the [cell’s] bigger machines.”
The groundwork for this bigger picture approach has already been laid, wrote Ran Chang, Ph.D., and Peter Jackson, Ph.D., of the Stanford University School of Medicine, in an editorial in Science that accompanied the three studies.
“For many cancers, there is an extensive catalog of genetic mutations,” they wrote. “But a consolidated map that organizes these mutations into pathways that drive tumor growth is missing.”
With funding from NCI’s Cancer Systems Biology Consortium, that consolidated map is precisely what the UCSF and UCSD researchers are hoping to help create with their Cancer Cell Map Initiative.
A linchpin of their research is a technology known as affinity purification–mass spectrometry, which they use to identify protein–protein interactions that include known cancer-related proteins. In meticulous fashion, the researchers systematically analyze mutations in genes and their impact on protein–protein interactions in cells and use advanced computational methods to parse the resulting data and generate the network maps.
These maps, Dr. Hughes said, “get us close to knowing what’s happening in a cell at any given time.”
Other studies have created network maps, Dr. Krogan noted, but they have been much more limited in scope.
“Nobody has done it to this scale,” he stressed. Integrating genomics, proteomics, and biochemistry with the mass spectrometry analyses and advanced computational tools, he added, “makes this a flagship set of studies, in my opinion.”
A Cozy Relationship with HER3
In the head and neck cancer study, the researchers first identified genes that previous studies have shown are commonly mutated in the most common form of head and neck cancers, called squamous cell carcinoma. They focused particular attention on PIK3CA because it’s the most commonly mutated gene in this type of head and neck cancer.
They analyzed two cell lines of head and neck cancer and a cell line made from healthy cells from the upper esophagus. In all, they identified 771 protein–protein interactions linked to these cancer-related genes in the head and neck cancer cells and healthy cells. Nearly 85% of the interactions had never been reported.
In cancer cells with the most commonly mutated form of PIK3CA, the resulting PIK3CA proteins “preferentially interacted” with HER3, they reported.
Like HER2, which is the target of several drugs now commonly used to treat breast cancer, HER3 is a receptor that straddles the cell’s surface and interacts with molecules both inside and outside the cell. A tight bond between HER3 and PIK3CA was not found in cells with normal PIK3CA protein or with different mutant forms of PIK3CA.
When the researchers tested an experimental HER3-blocking drug in mice injected with head and neck cancer cells with that specific PI3KCA mutation, no tumors grew. When they used cells with a different PI3KCA mutation, the HER3-blocking drug had no effect on tumor growth.
It would have been extremely difficult to make this discovery using more traditional approaches, Dr. Krogan said.
“It’s only when you do this type of analysis systematically and quantitatively that you’re able to see” this interaction between these two proteins, he said.
Pushing Ahead on Pathways
Based on previous studies implicating HER3 in head and neck cancers positive for the human papillomavirus (HPV), the HER3 inhibitor used in the head and neck study is already being tested in small clinical trials of patients with these cancers. But the study team believes these new findings may help identify people whose tumors are most likely to shrink following HER3-inhibitor treatment.
There’s a strong interest among researchers to target the “particular axis” of HER3 and PIK3CA interactions, Dr. Krogan said. Approximately 5% of all cancers have the specific PIK3CA mutation shown in the study to be susceptible to blocking HER3, he said.
So it’s not just head and neck cancer, “it’s many others,” he said.
From a broader perspective, continued research is needed “to further improve functional maps and more precisely define which [protein–protein] interactions are rewired in each tumor,” Drs. Chang and Jackson wrote in their commentary.
That will entail expanding the research scope, they stressed.
When proteins interact with each other, the resulting complexes “are not static structures, but rather dynamically reassemble in response to the state of the cell,” they explained. So it will be helpful to understand the impact on these interactions of other factors that can affect cells, they wrote, such as things happening in and around the tumor (known as its microenvironment), including inflammation and the presence of different types of immune cells.
Dr. Hughes agreed that these first studies are just the beginning. In the longer term, she anticipates that the development of tests that identify specific interactions in patients’ tumor samples would be difficult. Rather, she continued, “we’ll use the maps constructed through these types of studies to direct treatment by interpreting the genomic data on tumors already being collected.”
In the meantime, Dr. Ideker said that he’s optimistic that their work will encourage other groups to use this approach to mapping network pathways “and make it systematic for the whole of cancer cell biology.”